We gene edit several different strains of mice, including C57BL/6, Balb/c, DBA2, and NOD-scid for use as disease models. Mouse model generation can use either CRISPR/CAS9 EGE™ or ESC-based gene editing technologies, for which the turnaround times are 6-8 months and 9-12 months, respectively. Mice are often the preferred mammalian model organisms due to their short life cycle and close genetic resemblance to humans. Particularly, humanized mouse models have great potential. Mice happen to be the best studied mammalian model organism with abundant genomic, transcriptomic, proteomic, and phenotypic collections.
Strains
Laboratory mouse strains are either outbred or inbred. Inbred strains (also called inbred lines) are particular species that have individual mice nearly identical to each other in genotype due to long inbreeding. A strain is inbred when it has undergone at least 20 generations of brother x sister or offspring x parent mating, at which point at least 98.6% of the loci in an individual of the strain will be homozygous, and each individual can be treated effectively as clones. C57BL/6, BALB/c, Fvb, A/J, C3H, CBA, DBA/2 etc. are inbred mouse strains.
Application Example
Humanized CD3e Mouse Model Generation and Validation
Gene Editing Strategy
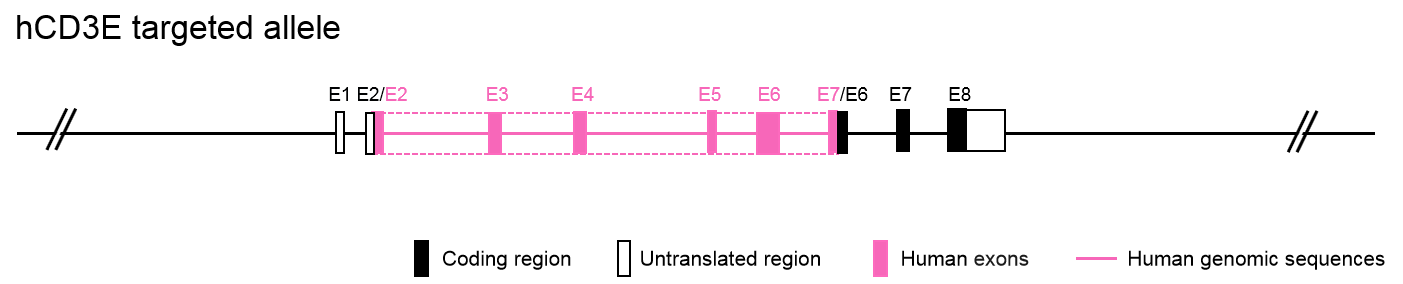
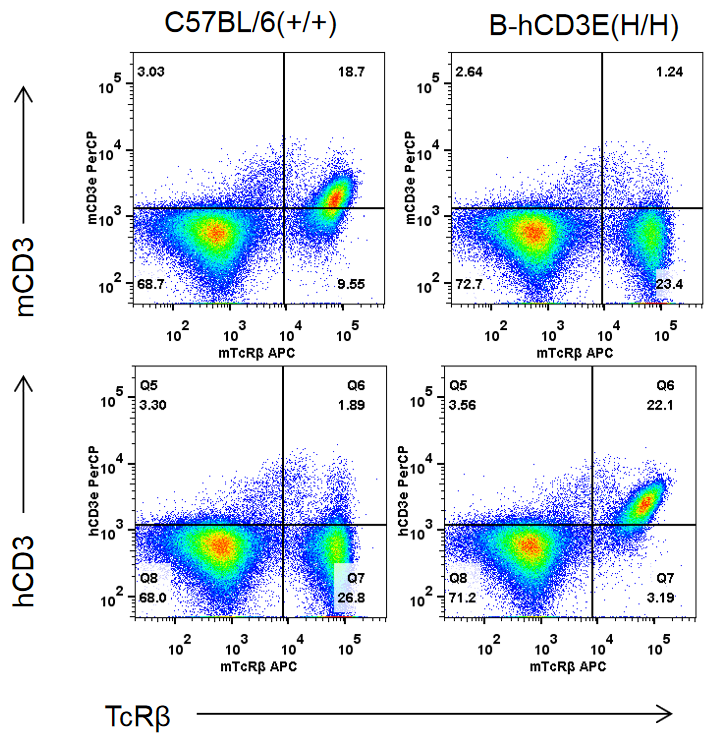
mRNA Expression Analysis

Protein Expression Analysis
Among the most commonly used mammalian experimental animal models, rat models offer many advantages over the mouse and other organisms.
The physiology of diseased rat models not only resembles more closely to that of humans but it is also comparatively easier to monitor in rats than mice. Popular experimental rat model strains include Sprague Dawley and Wistar. Biocytogen's transgenic rat service uses CRISPR/Cas9-based EGE™ to generate gene knockout and knockin rat models.
Application Example
The First Reporter Knockin Rat Developed by Biocytogen in 2014
As shown below, a pCAG-loxP-3X STOP-loxP-tdTomato-WPRE-bGHpA cassette is inserted between the first and second exons of the Rosa26 site using EGE™ technology. After mating cKI rats with Crh-Cre rats, tdTomato is highly expressed in the hippocampus of the Crh-Cre/tdTomato offspring.
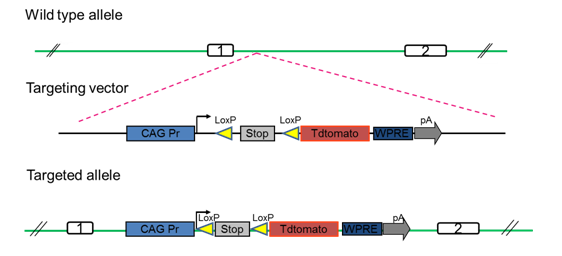
Targeting Strategy
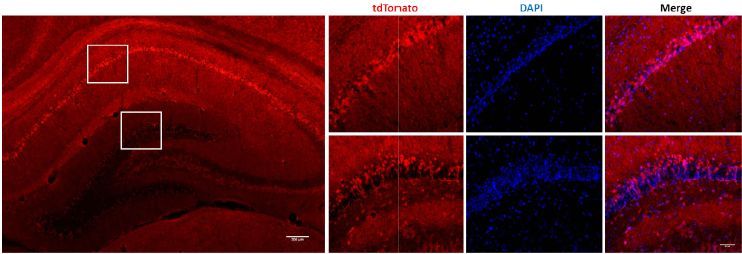
Histofluorescence experiments on the offspring of TdTomato rats mated with brain-specific-Cre-expressing rats
Although gene edited animal models are very powerful tools for biomedical research, genetically modified cell lines have many advantages including convenience, low cost, and adaptability for high-throughput screening. Biocytogen cell line gene editing services include feasibility evaluation, protocol design, cell transfection, single cell cloning, genotyping and validation, and any further necessary phenotypic validation study.
Application Example 1
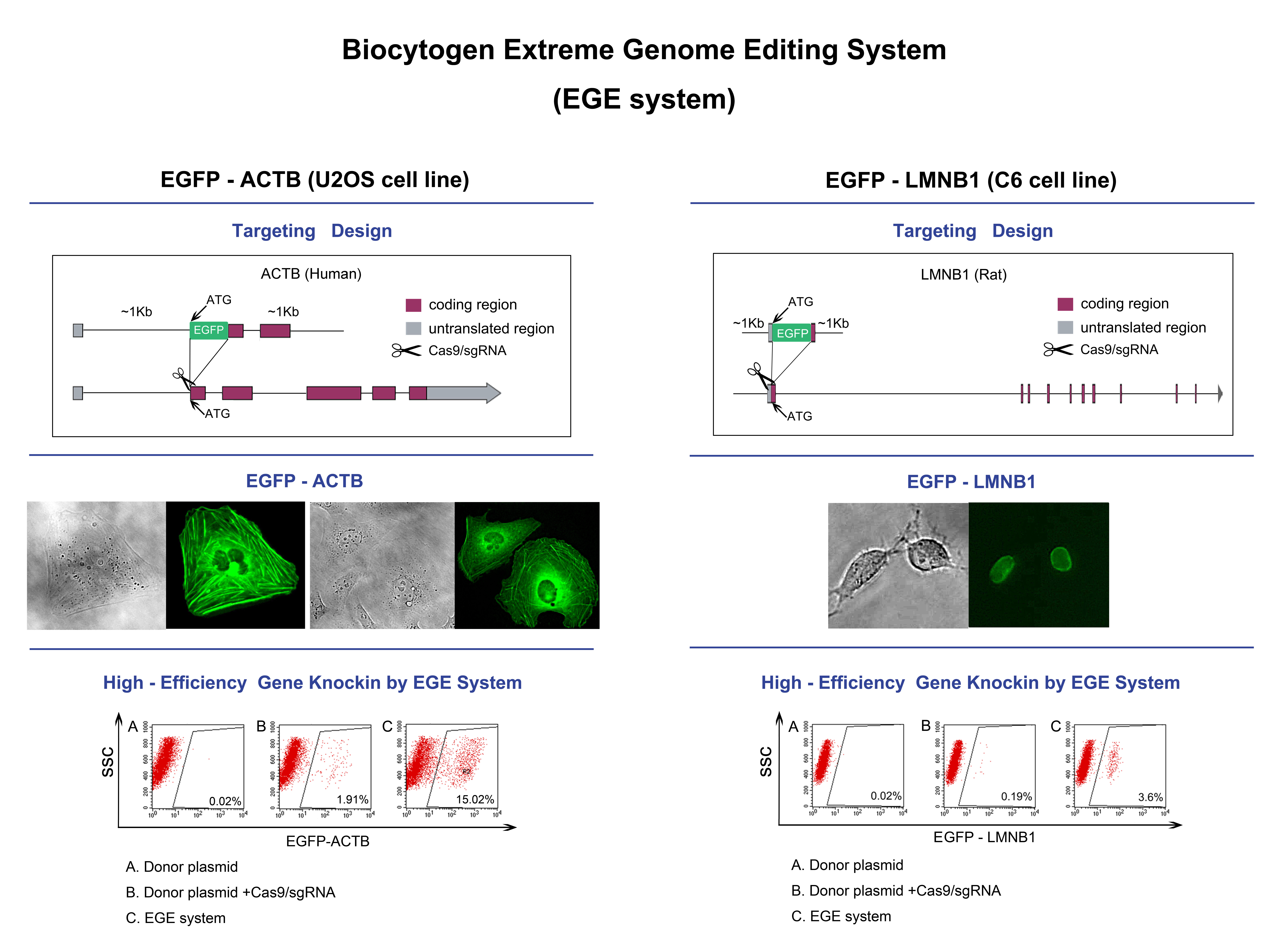
A knockin cell line model was generated with EGFP inserted at the translation initiation site of ACTB (cytoskeleton filiform protein) in the U2OS cell line (human osteosarcoma).
ACTB gene encodes β-actin, a cytoskeletal protein. The homologous arms of the targeting vector are about 1 kb, and the homologous recombination of the target vector was done by CRISPR/cas9 based EGE™ technology.
A similar gene editing strategy was used for LMNB1, which encodes the nucleoprotein Lamin-B1.
In U2OS cells, flow cytometry plots show that the efficiency of EGFP-ACTB gene knockin mediated by EGE™ (15.02%) is 8 times higher compared to CRISPR/Cas9-mediated knockin (1.91%). In the C6 cell line, the EGE™ system improves the knockin efficiency of EGFP-LMNB1 to 3.6% from 0.19%, which is a 19-fold increase.
Application Example 2
Gene Editing of hESCs and iPSCs
The highly efficient EGE™ system is ideal for editing the genomes of induced pluripotent stem cells (iPSCs) and human embryonic stem cells (hESCs).
Multiple iPSC lines are reprogrammed from human fibroblasts by non-integrated plasmids. Panel A shows phase contrast images. This iPSC knockin cell line expresses the pluripotency markers OCT4, SSEA-4, and TRA-1-60 as shown in Panel B and have a normal karyotype (Panel C). Teratoma forms in immunodeficient mice (Panel D).






 +86-10-56967680
+86-10-56967680 info@bbctg.com.cn
info@bbctg.com.cn