The strategies that our customized models use include:
Note: Selection marker for gene targeting is generally not required for animal model generation using CRISPR/Cas9-based EGETM, in contrast to ESC/HR technology. The following strategies with resistance markers are based on the traditional ESC/HR technology, and those without such markers are based on EGETM.
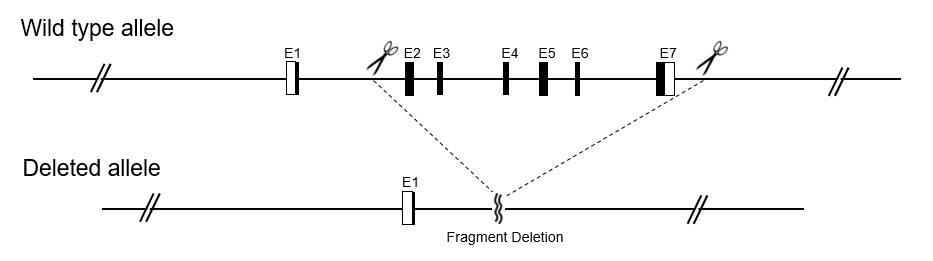
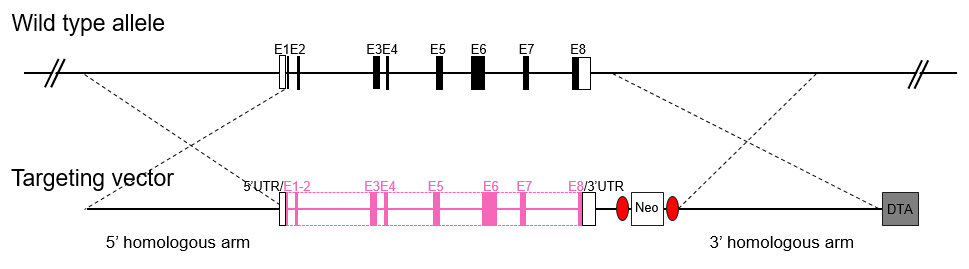
In a global knockout (KO) mouse model, and exon of a target gene is globally deleted (EGE™ method) or replaced (ESC/HR) with a positive selection marker, thus inactivating the gene. In global or whole-body KO mice, the gene of interest is disrupted in every tissue.
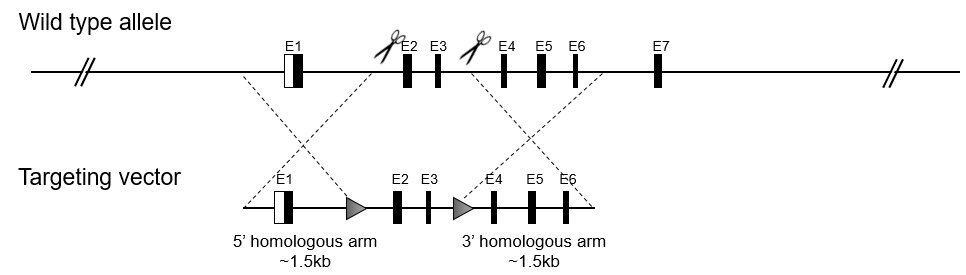
A conditional Knockout (cKO) model is generated via the Cre-LoxP recombination system. The targeted fragment to be knocked out is flanked by LoxP elements. Floxed mice are then bred with tissue-specific Cre mice, and sequences between LoxP sites will be removed from the offspring's genome in a tissue-specific pattern. Gene knockout is achieved by frame-shift after DNA sequence deletion.
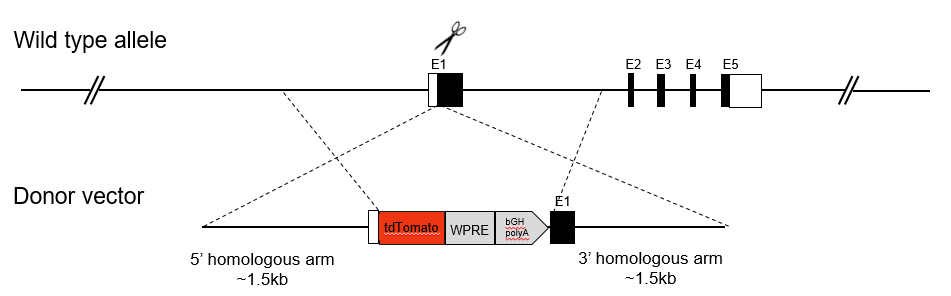
Global knockin mouse models introduce exogenous DNA sequences into a specific locus in the mouse genome. These models can mimic genetic diseases when mutations are introduced, or monitor gene expression when genes are tagged with various proteins (e.g. EGFP, mRFP, mCherry, YFP, LacZ, and Flag).
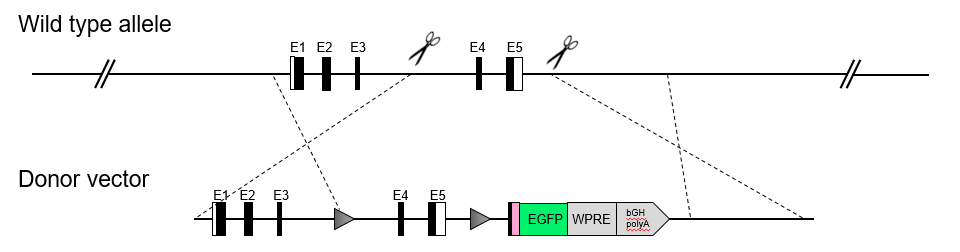
In a conditional knockin mouse model, mice containing a floxed (or conditional) allele are crossed with Cre-expressing mice, allowing for tissue-specific expression of any foreign gene.
The Cre recombinase protein mediates a tissue specific deletion of DNA fragments that are flanked by loxP sites. Therefore, the Cre mouse model allows tissue specific gene knockout based on the promoter driving Cre expression. The ERT2 sequence is an estrogen receptor targeting motif, whose nuclear translocation and activity are controlled by Tamoxifen. Thus, administration of Tamoxifen enables temporal or inducible control of Cre activity and gene deletion.
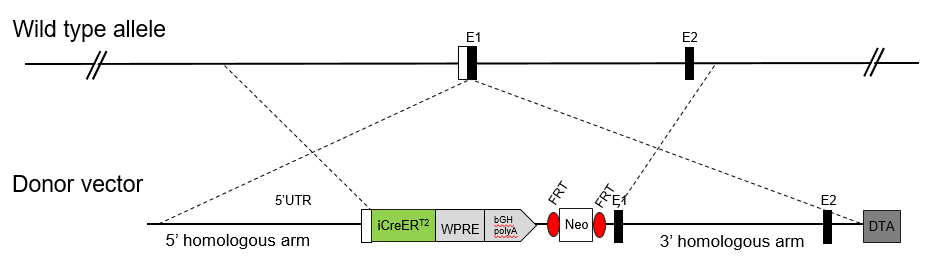
Conventional Point Mutation
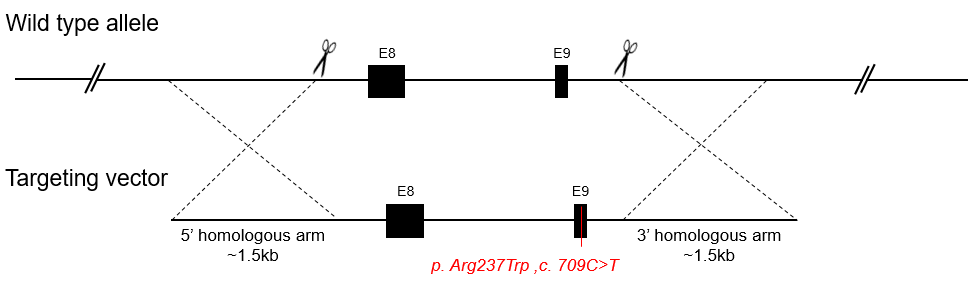
A conventional point mutation mouse model is a knockin mouse line in which one or more nucleotides in the mouse genome are substituted by variant nucleotides. This can result in an in-frame amino acid change of protein sequence. Point mutation knockin models are widely used to study the roles of particular nucleotides or amino acids within proteins, and to model human genetic diseases.
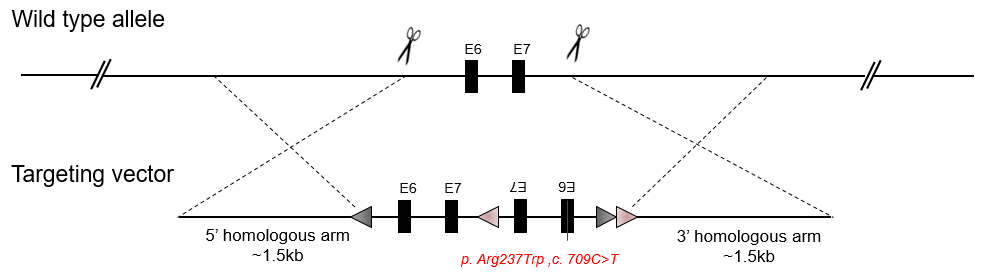
Conditional Point Mutation
A conditional point mutation mouse model introduces a point mutation when certain conditions are met. In the strategy below, when Cre recombinase is present, a point mutation will be introduced into the mouse genome tissue specifically via Cre activity. There are two major design strategies illustrated below:
Conditional Point KI by Double LoxP (Flex)
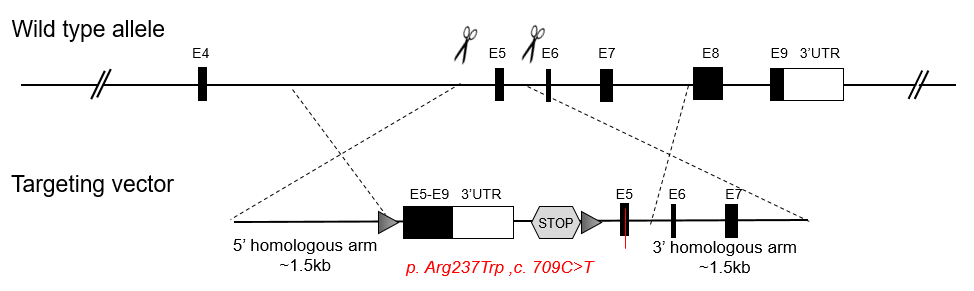
Conditional Point Knockin
Mouse-derived genes are knocked out and human-derived genes are knocked in to establish a humanized mouse model for pathogenetic and therapeutic studies of human disease models.
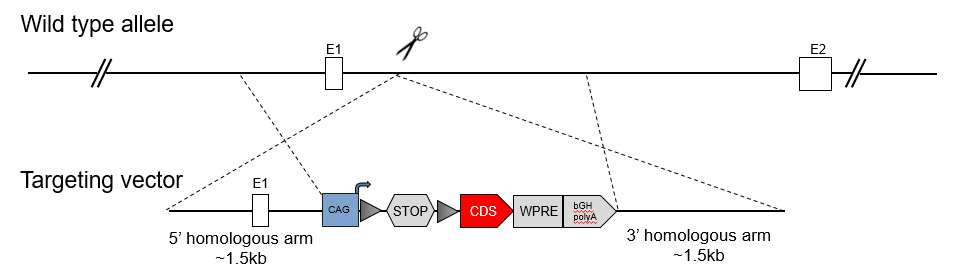
Traditional transgenic mouse models are usually constructed via pronuclear injection of plasmid, and many different founders can be obtained. The experimental results from different founder mice can be quite different and not reproducible due to the difference in integration copy number and loci. Currently, most investigators utilize site-specific integration strategies to construct gene-edited mouse models. Rosa26 is the most commonly used "safe harbor" locus because Rosa26 encodes a nonessential nuclear RNA expressed in almost all tissues. An exogenous gene will be conditionally expressed when a LoxP-STOP-LoxP sequence is inserted upstream of the exogenous sequence at the Rosa26 locus, and this model is crossed with a Cre deleter. There are also some other safe harbor loci, such as H11 and TIGRE.
Rosa26 Locus Knockin Model
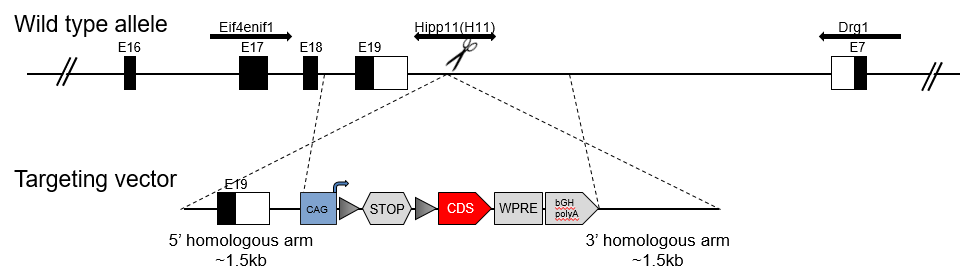
Hipp11 Locus Knockin Model
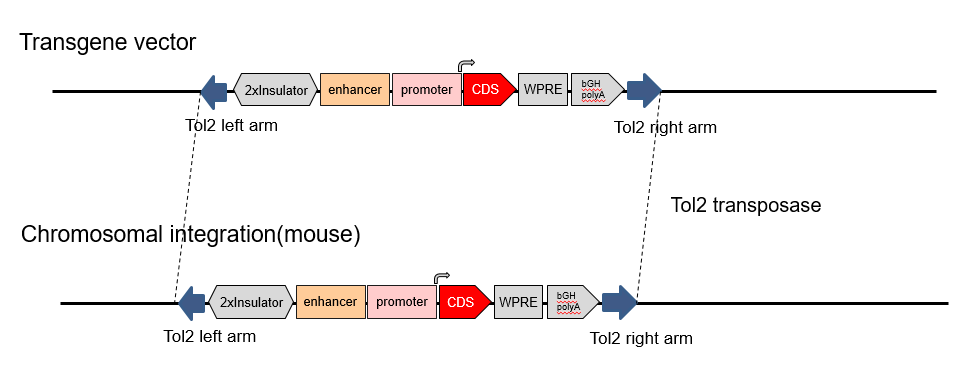
The Tol2 mouse model allows generation of transgenic mice utilizing Tol2 transposase activity. The Tol2 transposon system not only can increase the gene integration rate, but also has the inclination to integrate foreign genes into AT rich regions.






 +86-10-56967680
+86-10-56967680 info@bbctg.com.cn
info@bbctg.com.cn